近期在处理小鼠VBM数据,参考的是这篇文章的教程《Brain structure and synaptic protein expression alterations after antidepressant treatment in a Wistar–Kyoto rat model of depression》,其中内容为:The T2-weighted images analysis was performed using Statistical Parametric Mapping 12 (SPM12; https://www.fil.ion.ucl.ac.uk/spm/software/spm12/) and Data Processing & Analysis of Brain Imaging (DPABI) (Yan et al., 2016). First, each T2-weighted image was transformed from their original format of DICOM to NIFTI and re-sized voxels by a factor of 10 so that the rat brain volume will resemble the human brain and match the most parameters in SPM12. Then the rat brain was manually extracted from surrounding skull tissue and refined with a skull stripping tool in BrainSuite (http://brainsuite.org). After brain extraction, each rat brain was co-registered to SIGMA template (Barrière et al., 2019) and re-sampled into 1.5 mm voxels. Next, each image was segmented into three tissue priors (Gray Matter, White Matter, Cerebrospinal Fluid maps) using the old segment tool in SPM12. In this step, the default tissue probability maps were replaced by the SIGMA template’s GM, WM, and CSF tissue maps. To perform a more accurate analysis, all the segment priors were used to generate a subject-specific template by DARTEL (Diffeomorphic Anatomical Registration Through Exponentiated Lie algebra) algorithm in SPM12 as follows: 1 the “seg_sn.mat” documents generated during the old segment were used for initial import step; 2 GM, WM and CSF templates were created by DARTEL using imported three tissue priors respectively; 3 the Jacobian images acquired during DARTEL template creation of GM priors were applied to warp individual T2-weighted images; 4 the normalized T2-weighted images were averaged to generate final subject-specific template; 5 the GM priors were normalized to a subject-specific template and modulated. Finally, the normalized GM maps were smoothed using an 8-mm FWHM Gaussian kernel.
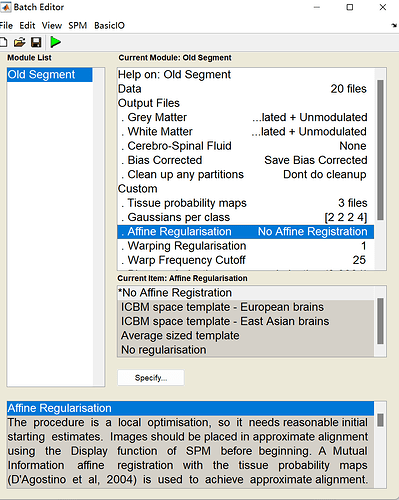
在进行old segment中,我不知道这里affine regularisation应该选择什么比较好,文章也没有写,而且发现用no affine registration和ICBM space template差别还挺大的,希望老师能给予意见,要是有懂小鼠VBM和DTI数据处理的,我可以给予付费咨询,也请联系,谢谢